Scientists working on a Centers for Disease Control and Prevention-funded project, led by Rice CS' Todd Treangen, has compiled a list of mutations in the SARS-CoV-2 spike protein that may make the omicron variant more resistant to neutralizing antibodies, including those from vaccinations.
That could increase the risk of breakthrough cases of COVID-19. The first such breakthrough in a fully vaccinated patient, though without a booster, in the United States was reportedly found in California this week.
A blog post by Gene Godbold, a senior scientist at Signature Science LLC, lays out nine of the dozens of reported genetic mutations and deletions in the omicron variant’s spike that the researchers suspect help the virus avoid antibodies.
The data is bound for submission as a peer-reviewed publication, but the rapid spread of omicron around the world gives urgency to aggregating and sharing the findings while awaiting more data to assess the implications, said Rice computer scientist Todd Treangen, principal investigator of the study who brought Godbold and his company onboard.
Treangen has long specialized in solving the computational challenges presented by the current deluge of medical data, with special focus on tracking pandemics.
Godbold said the CDC project has allowed the team to aggregate a wealth of data being compiled about the virus.
“What we do is make sense of millions of SARS-CoV-2 genome sequences by combining literature citations and sequences in a way that makes it easy to interpret new viral variants soon after they are documented,” he said.
“These kinds of assessments could not be attempted without the hundreds of researchers around the world who have dedicated countless person-years to work on the virus to ascertain how the mutations function in both laboratory and clinical settings,” Godbold said.
While additional data is needed to assess the severity of COVID-19 cases from omicron infection, Godbold wrote that the variant is likely to make SARS-CoV-2 not only more transmissible but also “one that can evade neutralizing antibodies induced by vaccination better than previously seen variants.
“This is not to suggest that vaccination is likely to be defeated by the omicron variant, only that the protection conferred by vaccination is likely to be shakier than with previous variants of concern,” he wrote.
The finding is part of a project that began in July with the CDC’s grant to develop the Treangen lab’s Harvest Variants software for the integrated, collaborative variant tracking of SARS-CoV-2. Godbold and his colleagues at Signature Science were contracted to curate variants of the virus and test third-party software and documentation services.
Harvest software, introduced by Treangen in 2014, is intended to be a suite of open-source microbial core/genome alignment and visualization tools that enable real-time monitoring of viral mutations. The CDC grant allowed Treangen and his team to fast-track its adaptation to monitor SARS-CoV-2.
“Gene’s efforts have been tremendous since he began work on this CDC project back in July,” Treangen said, noting that his collaboration with Godbold and Signature Science goes back many years. “Gene had already been doing this kind of analysis for existing variants, poring through hundreds of papers that link positions on the spike to variants that are either more transmissible or evade antibodies. He has already carefully categorized hundreds of positions along the SARS-CoV-2 genome including in the spike protein, which is developing into a comprehensive resource the community can leverage for rapid characterization of a new variant of concern.”
Treangen said the Rice team’s goal is to build informatics and computational tools that make such rapid analysis possible.
“Since the SARS-CoV-2 virus will not stop mutating, our goal is to provide a community resource to rapidly identify, aggregate and share high-quality data that complements related efforts taking place across the globe,” he said.
The Treangen Lab has been working on research related to COVID-19 since summer 2020, when it received grants to study artificial intelligence applications to mitigation of the disease and to collaborate on the design of a novel SARS-CoV-2 test. Recently the lab took part in a University of Maryland study to quantify the amount of virus slipping through ill-fitting masks.
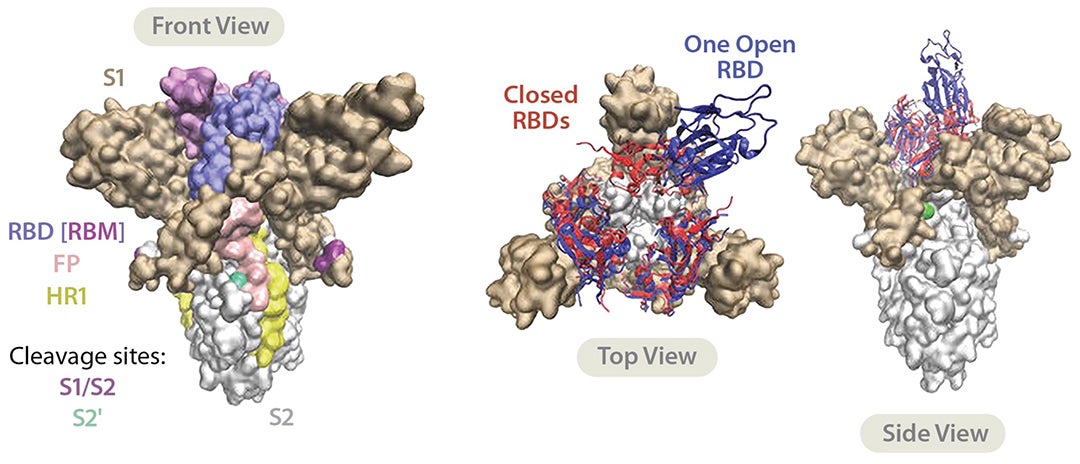
CAPTION: An illustration reproduced for the Signature Science blog with minor stylistic changes from the ScienceDirect article titled Peptide and peptide-based inhibitors of SARS by Desiree Schütz et. Al., published under a Creative Commons license.