When you take a medication, you want to know precisely what it does. Pharmaceutical companies go through extensive testing to ensure that you do.
With a new deep learning-based technique created at Rice University’s Brown School of Engineering, they may soon get a better handle on how drugs in development will perform in the human body.
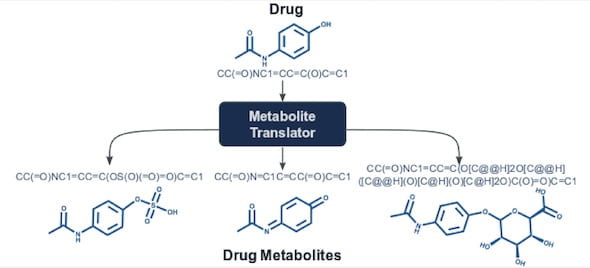
The Rice lab of computer scientist Lydia Kavraki has introduced Metabolite Translator, a computational tool that predicts metabolites, the products of interactions between small molecules like drugs and enzymes.
The Rice researchers take advantage of deep-learning methods and the availability of massive reaction datasets to give developers a broad picture of what a drug will do. The method is unconstrained by rules that companies use to determine metabolic reactions, opening a path to novel discoveries.
“When you’re trying to determine if a compound is a potential drug, you have to check for toxicity,” Kavraki said. “You want to confirm that it does what it should, but you also want to know what else might happen.”
The research by Kavraki, lead author and graduate student Eleni Litsa and Rice alumna Payel Das of IBM’s Thomas J. Watson Research Center, is detailed in the Royal Society of Chemistry journal Chemical Science.
The researchers trained Metabolite Translator to predict metabolites through any enzyme, but measured its success against the existing rules-based methods that are focused on the enzymes in the liver. These enzymes are responsible for detoxifying and eliminating xenobiotics, like drugs, pesticides and pollutants. However, metabolites can be formed through other enzymes as well.
“Our bodies are networks of chemical reactions,” Litsa said. “They have enzymes that act upon chemicals and may break or form bonds that change their structures into something that could be toxic, or cause other complications. Existing methodologies focus on the liver because most xenobiotic compounds are metabolized there. With our work, we’re trying to capture human metabolism in general.
“The safety of a drug does not depend only on the drug itself but also on the metabolites that can be formed when the drug is processed in the body,” Litsa said.
The rise of machine learning architectures that operate on structured data, such as chemical molecules, make the work possible, she said. Transformer was introduced in 2017 as a sequence translation method that has found wide use in language translation.
Metabolite Translator is based on SMILES (for “simplified molecular-input line-entry system”), a notation method that uses plain text rather than diagrams to represent chemical molecules.
“What we’re doing is exactly the same as translating a language, like English to German,” Litsa said.
Due to the lack of experimental data, the lab used transfer learning to develop Metabolite Translator. They first pre-trained a Transformer model on 900,000 known chemical reactions and then fine-tuned it with data on human metabolic transformations.
The researchers compared Metabolite Translator results with those from several other predictive techniques by analyzing known SMILES sequences of 65 drugs and 179 metabolizing enzymes. Though Metabolite Translator was trained on a general dataset not specific to drugs, it performed as well as commonly used rule-based methods that have been specifically developed for drugs. But it also identified enzymes that are not commonly involved in drug metabolism and were not found by existing methods.
“We have a system that can predict equally well with rule-based systems, and we didn’t put any rules in our system that require manual work and expert knowledge,” Kavraki said. “Using a machine learning-based method, we are training a system to understand human metabolism without the need for explicitly encoding this knowledge in the form of rules. This work would not have been possible two years ago.”
Kavraki is the Noah Harding Professor of Computer Science, a professor of bioengineering, mechanical engineering and electrical and computer engineering and director of Rice’s Ken Kennedy Institute. Rice University and the Cancer Prevention and Research Institute of Texas supported the research.