Doctoral Student Research in Computational Biology & Bioinformatics
Computational biology and bioinformatics research in the Department of Computer Science spans biological systems from individual genes, proteins, and cells, to networks of interacting molecules, to species and microbial communities. The research projects include protein structure prediction, protein binding and interaction networks, comparative genomics, microbial forensics, and phylogenomics. Research projects are conducted on a wide array of biological systems and species, including, but not limited to, cancer, neurodegenerative diseases (Alzheimer’s, Parkinson’s, etc.), microbiomes, and eukaryotic species.
The group members combine expertise and conduct research in computational disciplines as diverse as graph theory, algorithm design, combinatorial optimization, statistical inference, machine learning, and, more broadly, artificial intelligence. The group takes pride in implementing and making publicly available software tools for analyzing biological data and deriving hypotheses based on the new methodologies they design.
Faculty
Faculty members leading research in Computational Biology & Bioinformatics are as follows:
The videos below represent current student research projects.
|
|
Refined Protein Structural Prediction from 3D Patterson Maps via Recycling Training RunsPresenter: Evan Dramko
|
|
|
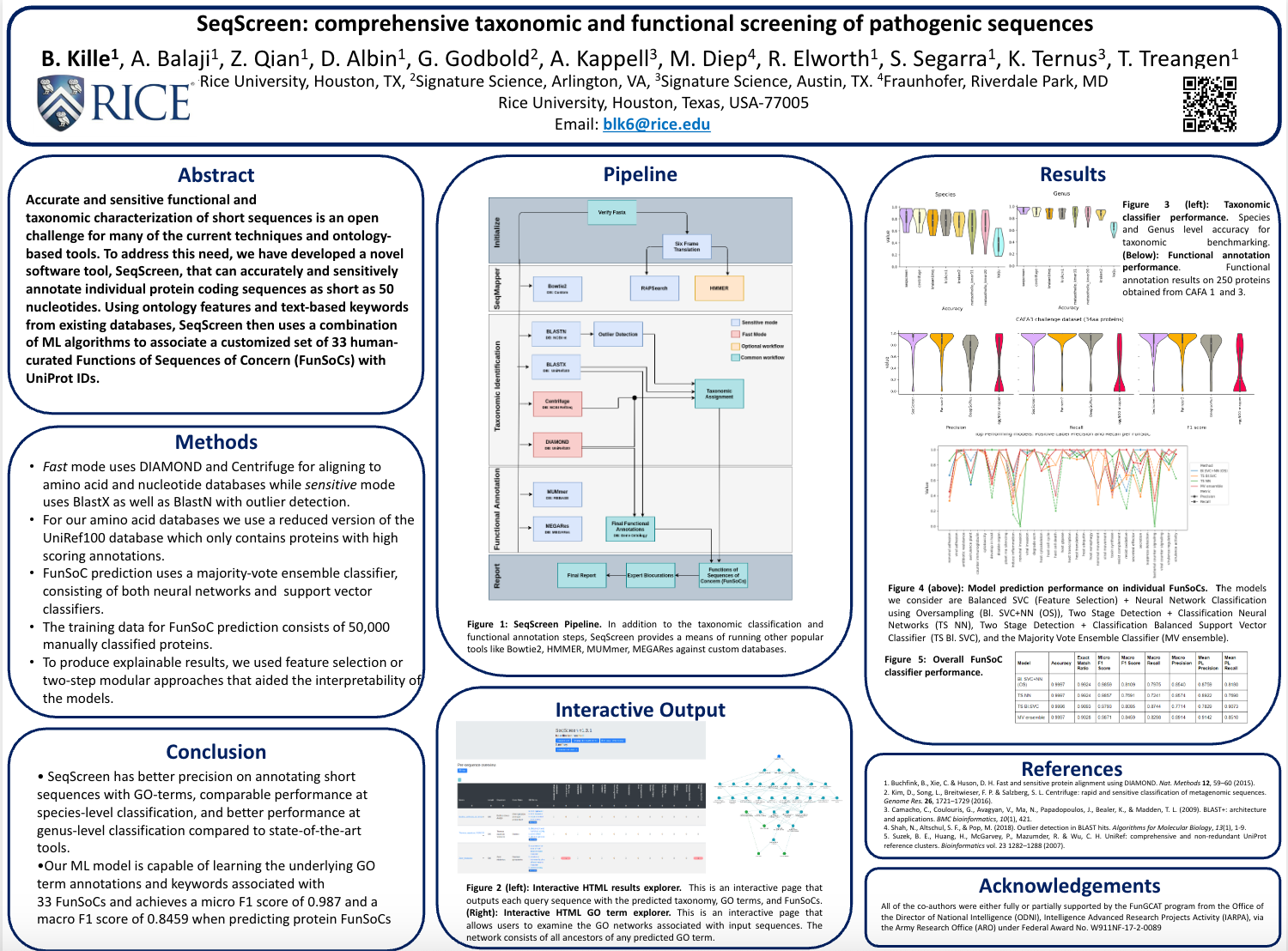
SeqScreen: accurate and sensitive functional screening of pathogenic sequences via ensemble learningPresenter: Felix Quintana [ Poster ]
|
|
|
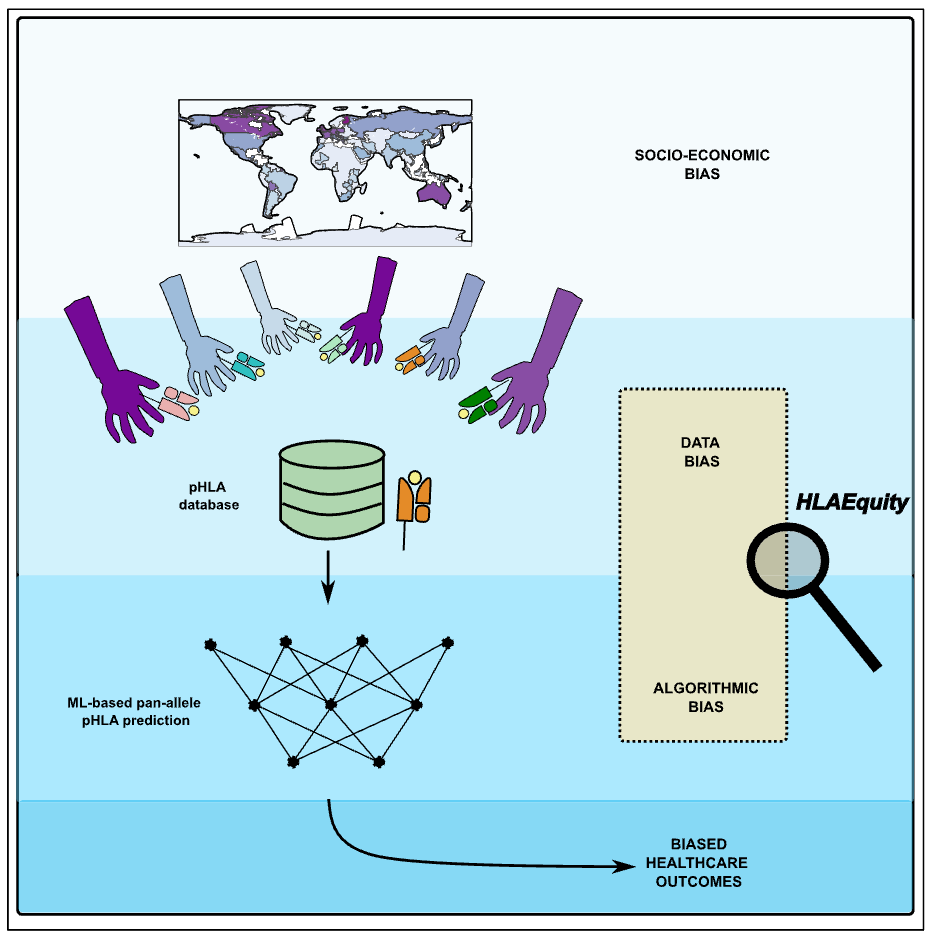
HLAEquity: Examining biases in pan-allele peptide-HLA binding predictorsPresenter: Alex Bock
|
|
|
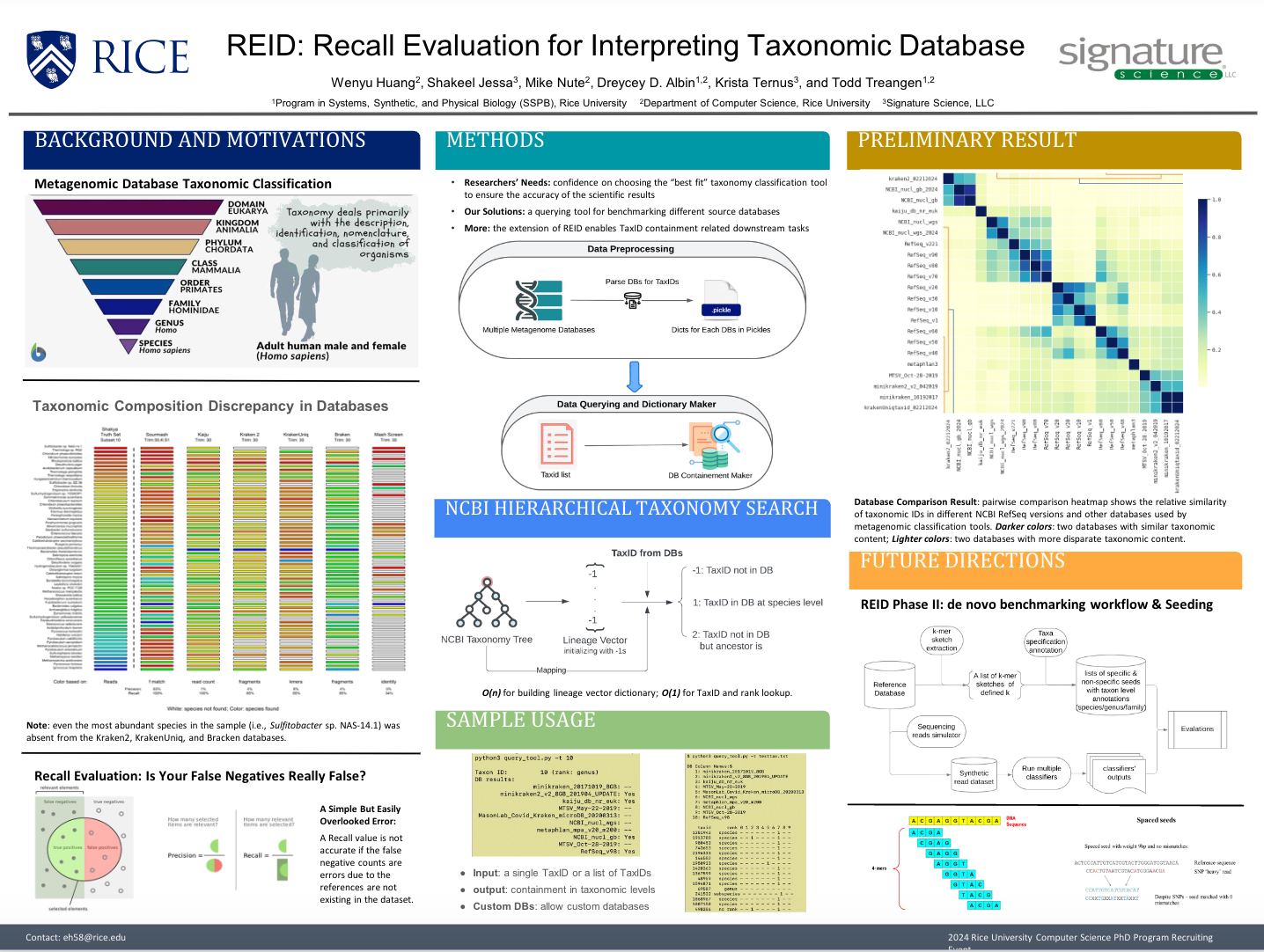
REID: Recall Evaluation for Interpreting Metagenomic Taxonomic DatabasesPresenter: Eddy Huang
|
|
|
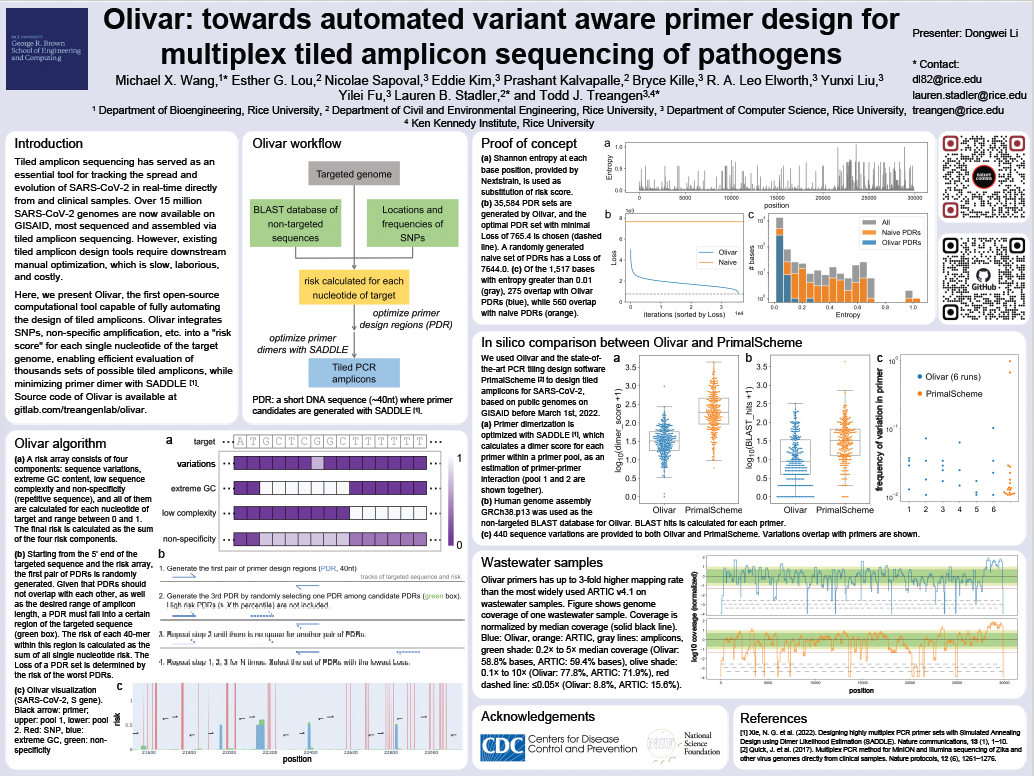
Olivar: Towards automated variant aware primer design for multiplex tiled amplicon sequencing of pathogensPresenter: Dongwei Li [ Poster ]
|
|
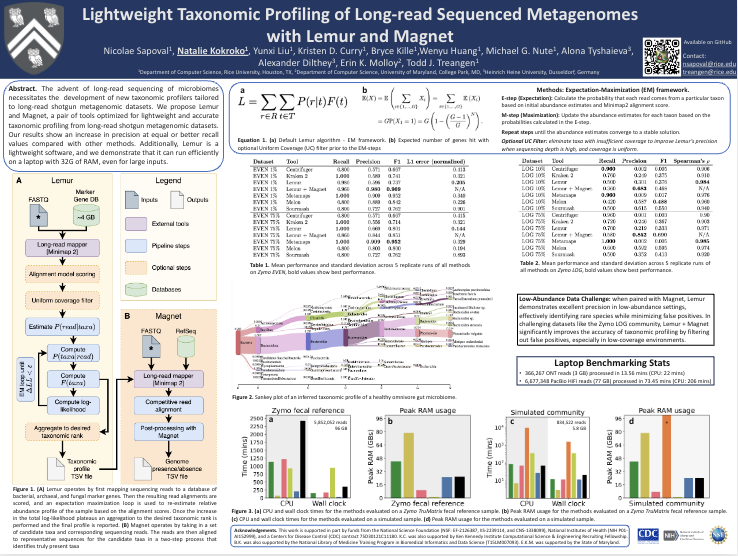
Lightweight Taxonomic Profiling of Long-read Sequenced Metagenomes with Lemur and MagnetPresenter: Natalie Kokroko
|
|
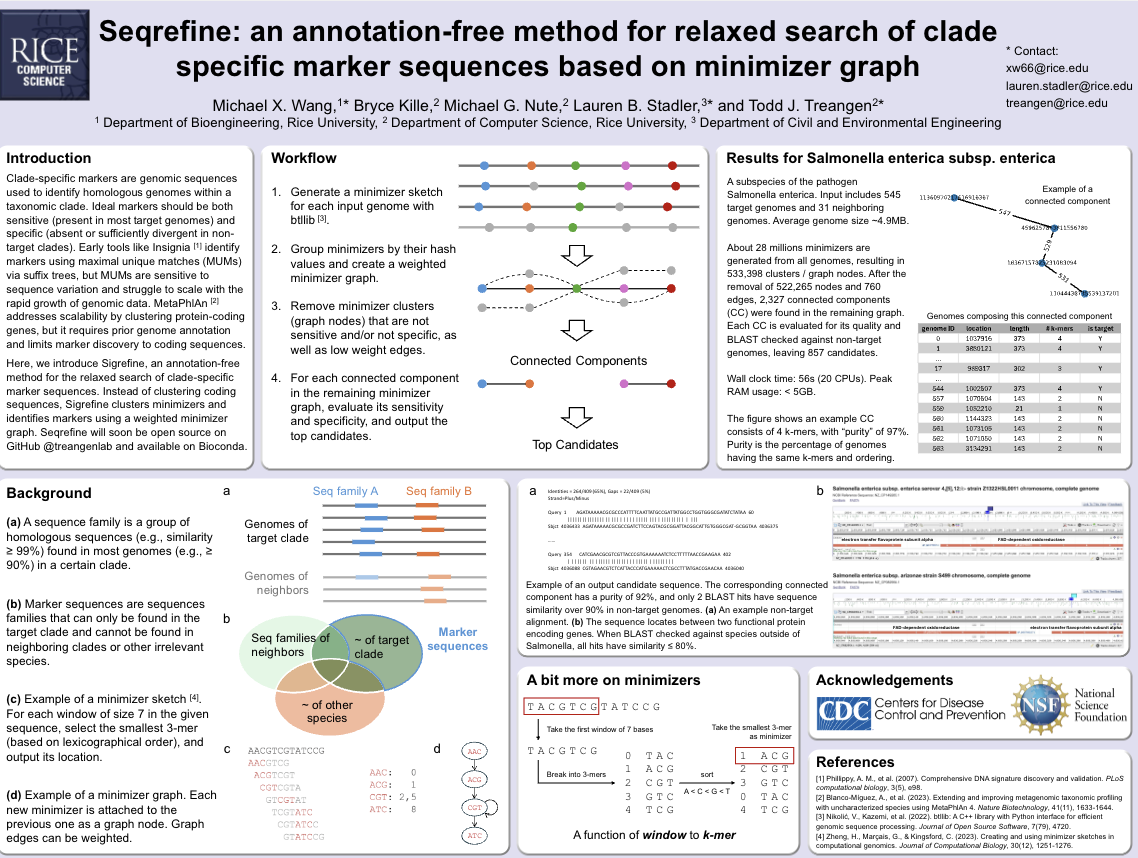
Clade specific marker region identificationPresenter: Michael Wang [ Poster ]
|
|
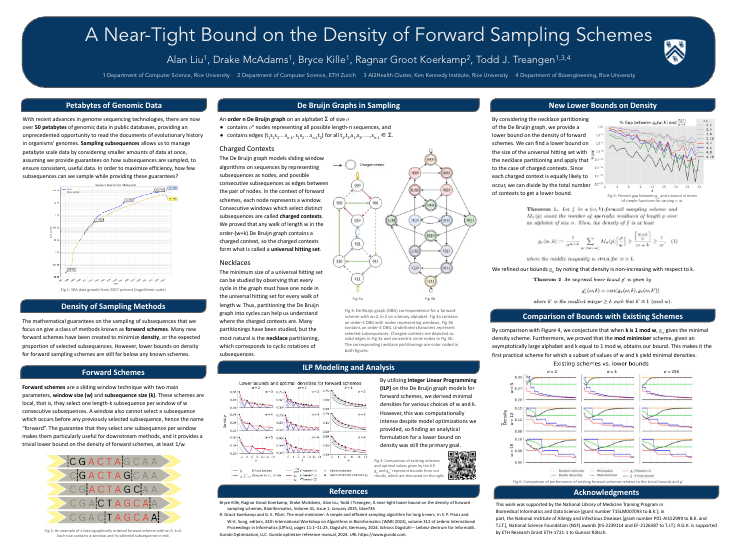
A near-tight lower bound on the density of forward sampling schemesPresenter: Drake McAdams [ Poster ] |
|
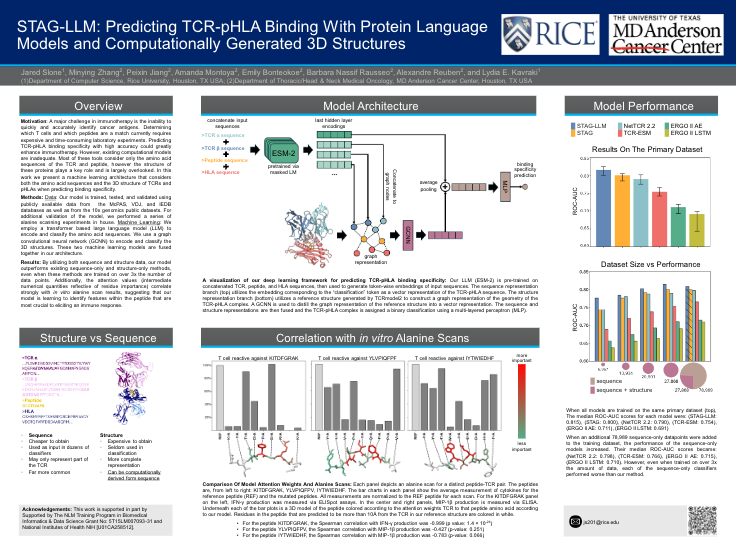
Utilizing protein language models and 3D structure to predict TCR-pHLA bindingPresenter: Jared Slone [ Poster ] |